You are using an out of date browser. It may not display this or other websites correctly.
You should upgrade or use an alternative browser.
You should upgrade or use an alternative browser.
News on China's scientific and technological development.
- Thread starter Quickie
- Start date
Important progress has been made in the field of DNA computing
Academician Fan Chunhai and Associate Professor Wang Fei from the School of Chemistry and Chemical Engineering/Transformative Molecular Frontier Science Center of Shanghai Jiao Tong University recently developed a DNA-based programmable gate array (DPGA) that supports universal digital computing, which can pass molecules The method of instruction programming realizes universal digital DNA computing and realizes the construction of large-scale liquid phase molecular circuits without attenuation. The results were recently published in the magazine Nature.
In 1994, Turing Award winner Adleman proposed using the complementary base pairing principle of DNA to develop biological computing. Since then, liquid-phase DNA molecular computing based on the interactions between DNA molecules has shown great potential in highly parallel coding and execution algorithms. Previously, researchers have used DNA molecular reaction networks to successfully implement multiple functions such as cellular automata, logic circuits, decision-making machines, and neural networks. However, the existing DNA computing system can only customize the hardware for specific functions. In the field of electronic computers, general-purpose integrated circuits (such as FPGA) can perform various computing functions through software programming without the need to design and manufacture hardware from scratch. This provides a high-end platform for the development of computing machines. Therefore, how to develop universal programming and integration of DNA computing components has become a bottleneck restricting the development of the field of DNA computing.
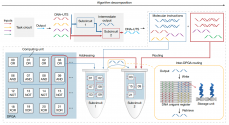
In response to this challenging problem, the research team first demonstrated that the use of single-stranded DNA as a unified transmission signal (DNA-UTS) can achieve a function similar to the transmission of electrons in a circuit. Furthermore, a DPGA was developed that supports general-purpose digital computing and supports multi-DPGA integration at the device level, achieving intra-device programmability and inter-device integration. As shown in (Figure 1), when the complexity of the circuit exceeds the executable scale of a single DPGA, the DPGA can be decomposed into multiple subtasks and generate corresponding molecular instructions; the molecular instructions of each subcircuit are called and connected through logical addresses to participate in the operation. DNA components realize DPGA programming; signal transmission between sub-circuits is realized through multi-DPGA wiring mediated by DNA origami registers, thereby achieving device-level multi-DPGA integration.
Is it actually possible to replace wifi with nearlink?Huawei debuts Nearlink a faster more secure alternative to Bluetooth.
I didn't really get the impression it was meant to replace wifi 6 maybe wifi direct. But the articles didn't went that deep.
I don’t think its meant to replace Wifi but rather its meant to replace Bluetooth.Is it actually possible to replace wifi with nearlink?
I didn't really get the impression it was meant to replace wifi 6 maybe wifi direct. But the articles didn't went that deep.
Chinese AI-developed drug offers hope to sufferers of obesity and diabetes
- Developed using artificial intelligence, the weight-loss and diabetes drug MDR-001 is currently in its phase 2 clinical trials
- The use of AI in drug discovery is helping to keep costs down and speed up development times of new treatments
With the help of artificial intelligence, Chinese have found a new weight-loss drug. And it can also treat type 2 diabetes.
The , developed by drug discovery company MindRank, has now entered its phase 2 clinical trials.
According to the Hangzhou-based company, the drug should also be able to hit shelves much faster than usual.
Is it actually possible to replace wifi with nearlink?
I didn't really get the impression it was meant to replace wifi 6 maybe wifi direct. But the articles didn't went that deep.
I don’t think its meant to replace Wifi but rather its meant to replace Bluetooth.
Nearlink is a set of two standards published by Sparklink Alliance (the standard body), namely Sparlink Low Energy (SLE) and Sparklink Basic (SLB). SLE is the counterpart to Bluetooth, SLB is counterpart to WIFI with technologies borrowed from 5G (the polar code by Huawei among others).
I don't know if the name Nearlink replaces Sparklink SLB and SLE all together, or only represents SLE. But the current implementation in Huawei products is certainly SLE standard. So the confusion is just a naming convention problem, not a problem/limit of technical specification.
Last edited:
SIOM has developed gratings for 50 petawatt-class high-power laser system that has an energy-loading performance of >7 times compared to the Lawrence Livermore Lab's National Ignition Facility's gratings developed in 2022. The PI is Shao Jianda who is the director of SIOM.
LLNL NIF leads the US inertial confinement fusion project. So they probably uses the same gratings in their laser based nuclear fusion.
LLNL NIF leads the US inertial confinement fusion project. So they probably uses the same gratings in their laser based nuclear fusion.
Last edited:
It says several times of BT, which is only a couple Mb/s, so not nearly enough bandwidth to replace wifi. Apparently it's enough for lossless audio though, which for audiophiles is absolutely a deciding factor over BT. You don't spend hundreds/thousands on DACs, Amps, and headphones just to listen to lossy audio like aptx, which is why high end headphones are still wired. If NearLink can do lossless, even if compressed lossless audio wirelessly then it could take over the whole high end market.Wow, 1/30 microsecond lag .... imagine that for WiFi! I wonder what the max bandwidth per second is for NearLink.
